
GSoC Gupta Lab
Biospectrogram
Biospectrogam is an open-source software for the spectral analysis of DNA and protein sequences [1]. The software can fetch (from NCBI server), import and manage biological data. One can analyze the data using Digital Signal Processing (DSP) techniques since the software allows the user to convert the symbolic data into numerical data using 23 popular encodings and then apply popular transformations such as Fast Fourier Transform (FFT) etc. and export it. The ability of exporting (both encoding files and transform files) as a MATLAB .m file gives the user an option to apply variety of techniques of DSP. User can also do window analysis (both sliding in forward and backward directions and stagnant) with different size windows and search for meaningful spectral pattern with the help of exported MATLAB file in a dynamic manner by choosing time delay in the plot using Biospectrogram. Random encodings and user choice encoding allows software to search for many possibilities in spectral space.
Software Homepage 
(Click the logo)
Prerequisites: Signal processing, spectral analysis, biological signal processing,
signal transformations, encoding techniques.
(Student without prerequisites can seek the knowledge in project understanding phase.)
Programming Skills: Matlab, java.
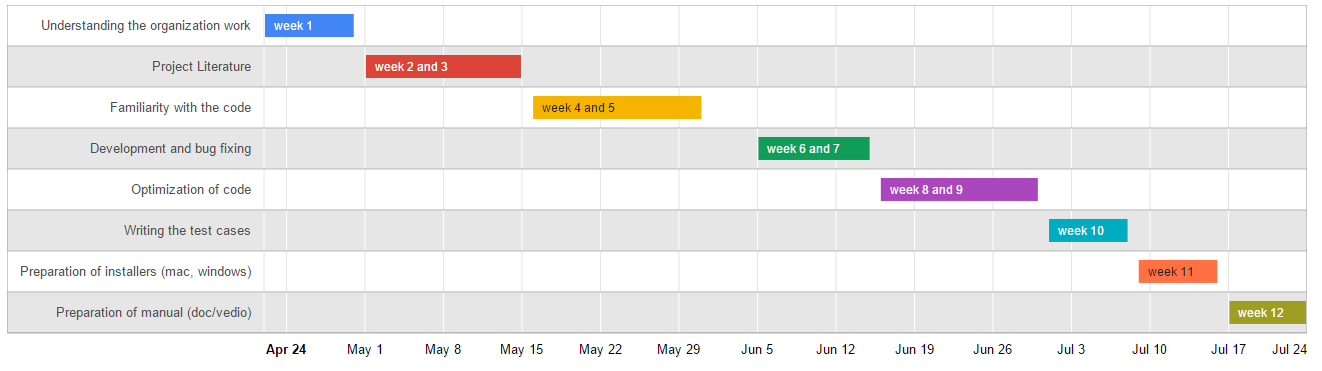
Road Map:
References:
1. Turakhia, Naman, et al. "Biospectrogram: a tool for spectral analysis of biological sequences." arXiv preprint arXiv:1210.1472 (2012).
Graduate Mentor
Dixita Limbachiya
Key Developers
1. Nilay Chedda (Master student in Computer science at University of Southern California)
2. Naman Turakhia (GPU Architect at NVIDIA)
Copyright © 2015- Dixita Limbachiya
signal transformations, encoding techniques.
2. Naman Turakhia (GPU Architect at NVIDIA)